Microplate reader uses "non-invasive" technology to detect live cell fluorescent protein
Introduction
In the past five years, fluorescent proteins have played an increasingly important role in monitoring in vivo biological research. Green fluorescent protein (GFP) from Victoria multi-tube luminescent jellyfish is the first fluorescent protein we have applied, but over time, the types of fluorescent proteins we can use now are more abundant, including enhanced variant GFP. Protein, fluorescent proteins and coral reef proteins found in other species of jellyfish. They can all be cloned and replicated in numerous cells and tissues, from bacteria to yeast, from plants to mammals. These fluorescent proteins are stable, have low cytotoxicity, and produce visible fluorescence in the body without additional assistance. Thus, they can be used as molecular targets or as independent reporters to visualize, track and quantify a variety of different cellular processes, including cell synthesis and metabolism, protein transport, gene induction, and cell lineage. Different fluorescent proteins have different colors and can therefore be used for multiplex detection. These fluorescent proteins can be detected by fluorescence microscopy and flow cytometry. But correspondingly, if we don't do cell sorting or monitor intracellular migration, the microplate fluorescence plate reader will be a better, more convenient, and more throughput detection system. Below, we will show you how the Molecular Devices microplate reader uses "non-invasive" technology to detect live cell fluorescent proteins.
We performed stable transfection of different fluorescent proteins from three HEK-293 cell lines obtained from BD Biosciences Clontech. The purpose of this study was to: 1) optimize the wavelength setting of the three cell lines, 2) determine the lower limit of detection LLD by diluting the cell line, and 3) demonstrate the feasibility of identifying another cell line in one cell line.
material:
The HEK-293 cell line stably expresses three fluorescent proteins:
AcGFP---another GFP produced in multi-tube luminescent jellyfish (but different from Victoria jellyfish)
ZsGreen---similar to GFP, but brighter (from coral reef)
DsRed--a red fluorescent protein non-transfected HEK-293 cell line from coral reefs as a control
method:
Cell preparation and analysis
The cells were cultured in a large flask, and a medium containing DMEM containing 10% FBS + 1% Pen/Strep/L-glutamine + 500 μg/mL G 418 was used. HEK-297 cells that were not transfected served as controls. One night before the experiment, the cells were trypsinized and then subjected to density gradient dilution, with a final density ranging from 500,000 to 100 cells per milliliter. They were then inoculated overnight into 96 well (100 ul) and 384 well (25 ul) plates. Therefore, the density of cells seeded is 50,000 to 10 cells per well (96-well plates) and 12,500 to 2.5 cells per well (384-well plates). Make 12 repetitions for each dilution. On the second day, the instrument was subjected to bottom reading and top reading using SpectraMax M5 and Gemini EM.
Wavelength optimization
The SpectraMax M5 and Gemini EM are both monochromator-based scanning fluorescence readers. For each particular fluorescent dye, both the excitation and emission wavelengths can be optimized over the background, which is not available with filter-type microplate readers. In general, the optimization strategy is: excitation with excitation light below the theoretical maximum excitation wavelength of 20-25 nm, and then initial emission of the emission wavelength; secondly, excitation of the excitation wavelength with an emission of light higher than the theoretical maximum emission wavelength of 20-25 nm. The actual excitation and emission wavelengths will be given after scanning. Finally, additional signal scans can be combined with the signal/background analysis to determine the final result. (According to Stockton's displacement theory, the excitation wavelength should be lower, the emission wavelength should be higher, and an emission light blocking filter is needed to isolate the non-target wavelength optical signal). And there may be two more scans because you want to choose the best barrier filter.
result:
Wavelength optimization
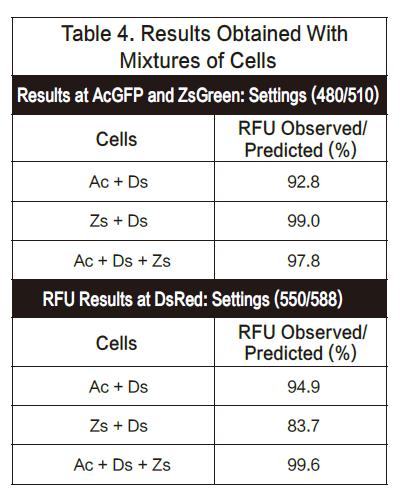
Wavelength scanning of DsRed with Gemini EM provides an example of how to optimize wavelengths. To detect the maximum excitation wavelength, we first fixed the emission wavelength to 600 nm and then scanned the emission wavelength. The scan results showed a maximum excitation wavelength of 556 nm. (Fig. 1) Also to detect the maximum emission wavelength, we fixed the excitation wavelength to 535 nm and then scanned the emission wavelength to obtain a result of 584 nm. (Figure 2) Both the excitation and emission values ​​are similar to the published wavelength results 557/579.
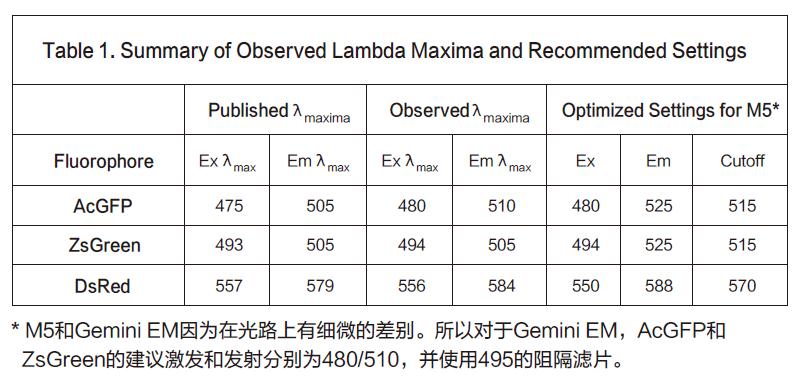
The next step is to determine the optimal excitation and emission wavelengths in conjunction with the signal/background optimization results. Because the Stoketon shift of the preliminary test results is small (22nm), it is obvious to increase the difference between the two by reducing the excitation wavelength and increasing the emission wavelength. Secondly, it is necessary to find a suitable emission light barrier filter to optimize the most. Good sensitivity. Finally, we used an excitation wavelength of 550 nm to excite while using an 570 nm emission blocking filter to isolate unwanted excitation light above 570 nm. DsRed transfected cells were scanned by an emission wavelength of 575 to 600 nm, and the results showed a peak appeared at 587-588 nm (Fig. 3, upper curve). In the background (cells that are not transfected), the spots are relatively flat (Figure 3, lower curve). Based on this scan, our final optimized settings were 550 nm excitation, 588 nm emission, and 575 nm as the emission light blocking filter.
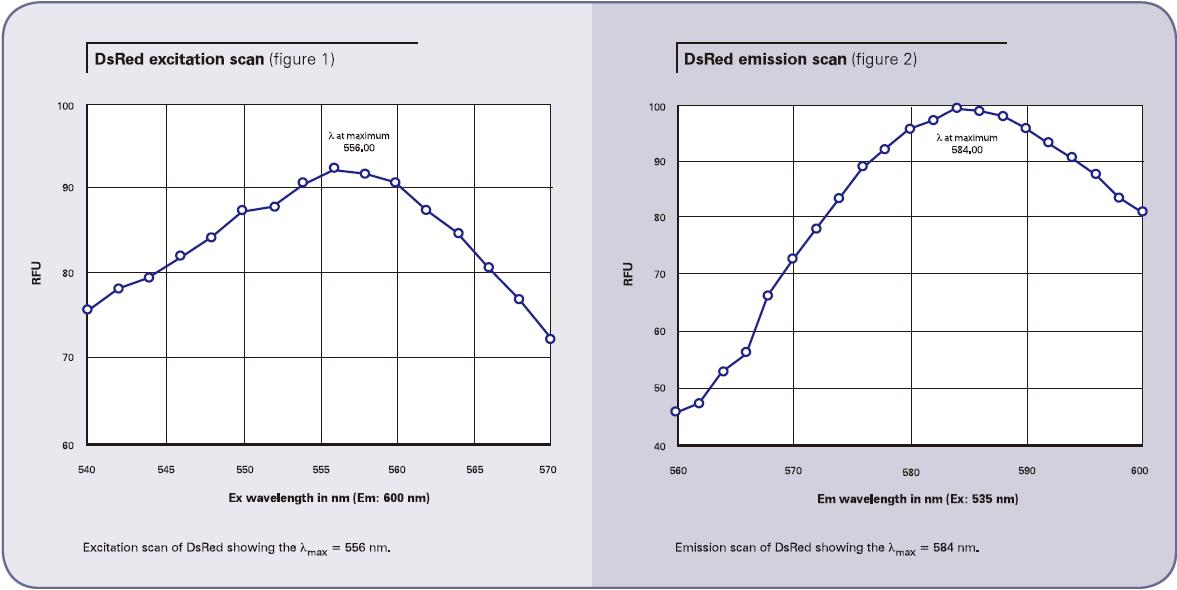
The maximum values ​​observed and the settings we recommend for qualitative analysis are summarized in Listing 1.
Cell dilution results
The cell dilution results obtained using the bottom reading are shown in Figure 4 (96-well plate) and Figure 5 (384-well plate). The top reading results are shown in Figure 6 (96-well plate) and Figure 7 (384-well plate). The brightness of the ZsGreen cell line (curve above) is nearly 3.5 times that of the other two cell lines. Although the results of the DsRed cell line were darker than ZsGreen, the detection sensitivities of the two cell lines were similar. (Bottom of Listing 2) because the background value is lower than the wavelength of DsRed.
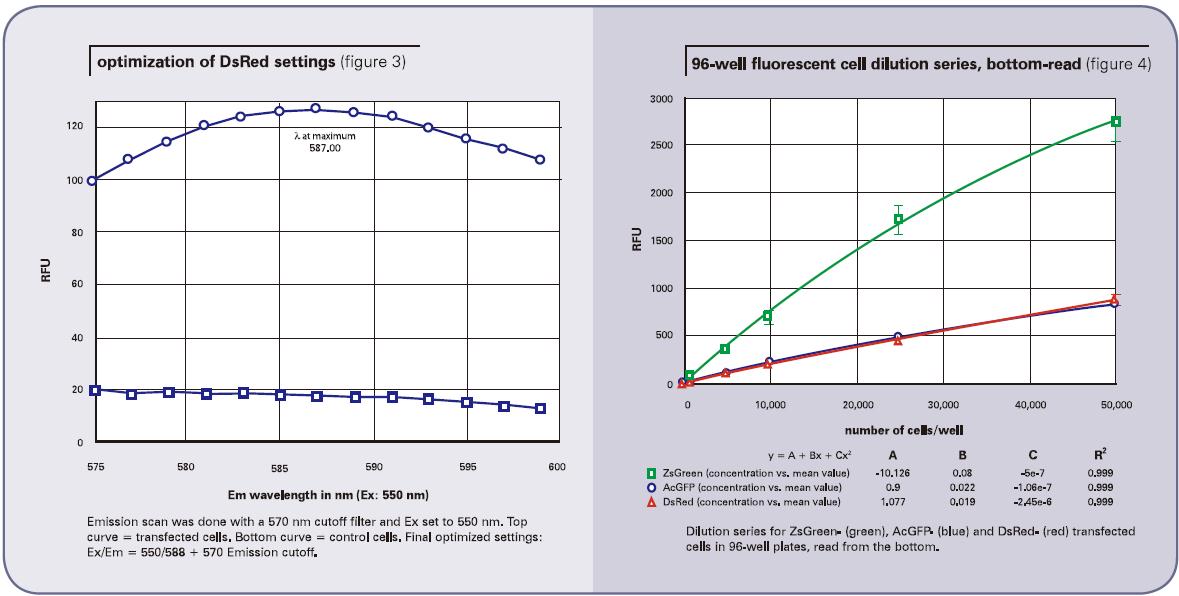
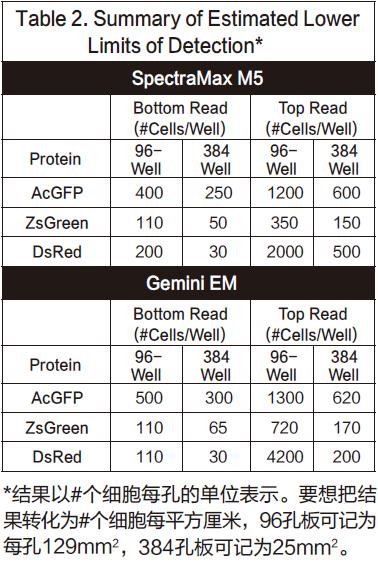
In general, the points in the figure are slightly non-linear, with a slight drop at the top. We conclude that because cells tend to adhere to the wall at high concentrations, they can cause non-linear results. Listing 2 gives the lower limit of detection LLD confirmed by all cell lines. The calculation method of LLD is: 3SDBLANK/Slope, SDBLANK is the standard deviation of background holes (medium without cells), and Slope is the slope at the bottom of the curve. (For 96-well and 384-well plates, data for 10,000 cells per well and 2500 cells per well were used). The RFU signal and standard deviation results for non-transfected cells were similar to those for media alone.
In this experiment, both the ZsGreen and DsRed cell lines have similar detection limits in the bottom-reading mode, and both are three to four times lower than the lower limit of detection of the AcGFP cell line. The experiment was repeated a total of three times, but the results of the DsRed experiment did not always perform well enough. In one experiment, its detection limit was similar to AcGFP, but in another experiment, its lower detection limit was high. We attribute these differences to the number of channels in different cell lines. (The darker the cell becomes with a successful channel.) The DsRed cell line grows faster than other cell lines at the beginning of the experiment, so in the first experiment it was 2-4 times more than other cell lines. The result of the channel is obviously not ideal.
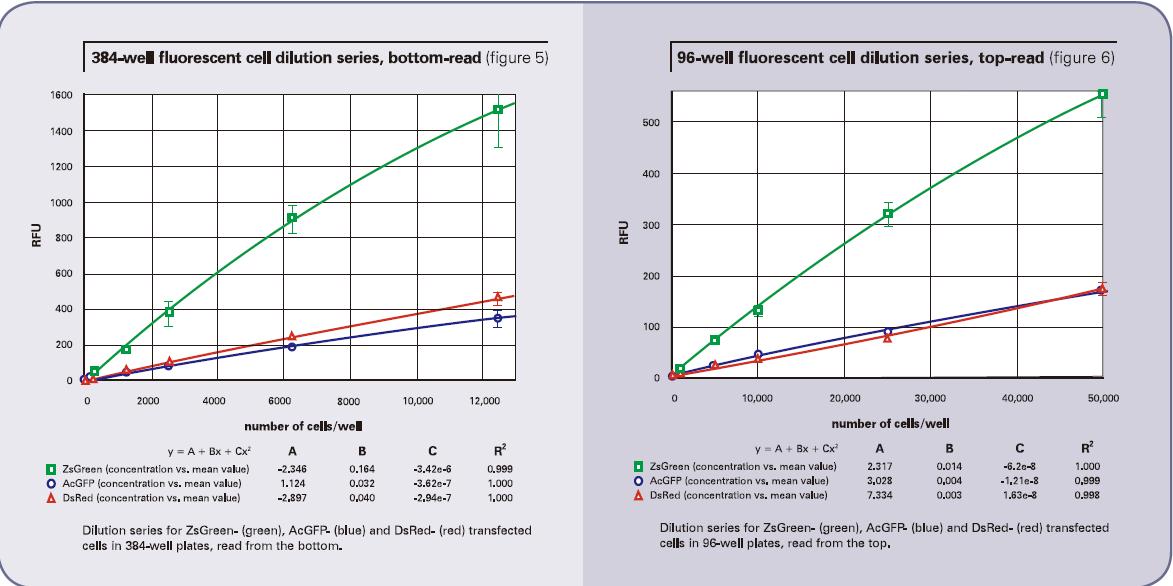
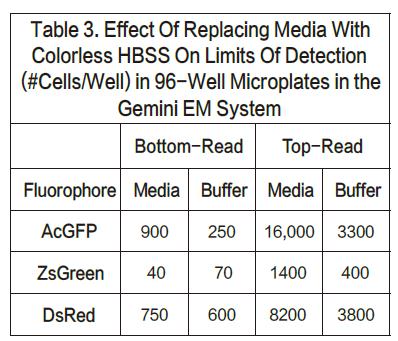
For the AcGFP and ZsGreen cell lines, the upper limit of detection was nearly three times higher when using top reading. On the other hand, for DsRed, when top reading is used, the detection limit will increase by nearly 10-20 times. We believe that the reason for this decrease in sensitivity is that the DMEM medium has a red fluorescent signal that has internal interference with the results. It has also been proven that by changing the medium to a colorless HBSS, the corresponding DsRed top reading results will also be better. Listing 3 shows the results of replacing the colored medium with a colorless medium. There was not much change in the bottom reading (although the lower detection limit of AcGFP appeared to improve), demonstrating that this method of operation did not result in cell loss. However, the top reading results in a nearly threefold improvement in sensitivity. These results also support a theory in one aspect that the colored medium will affect the sensitivity of the top reading to some extent.
Cell mixing result
In this experiment, we mixed a variety of cells in a 96-well plate to ensure approximately 50,000 cells per well. Therefore, if it is a 1:1 blend, there will be 25,000 per cell. If it is a 1:1:1 blend, then there will be 16 and 700 of each cell. The assay plates were tested with excitation and emission of 480/510 nm (optimized results for AsGFP and ZsGreen) and 550/588 nm (DsRed settings), respectively. The observed RFU values ​​are close to the predicted values. The results are summarized below. The results show that AsGF P and ZsGreen can be detected together with DsRed and vice versa. (See Figure 4)
in conclusion
Molecular Devices' fluorescence plate readers detect fluorescent proteins in intact adherent cells. The wavelength-scanning feature optimizes the wavelength results for each unique fluorescent dye. The bottom reading mode provides the best detection sensitivity, and the top reading mode also provides usable data, especially for cell lines containing ZsGreen and DsRed. When the DsRed cell line is detected by the top read mode, the purpose of optimizing the experimental results can be achieved by replacing the leuco medium.
Food flavoring
China Food flavoring Suppliers
Here you can find the related products in Food flavoring, we are professional manufacturer of Food flavoring Raw Material. We focused on international export product development, production and sales. We have improved quality control processes of Food flavoring to ensure each export qualified product.
If you want to know more about the products in Food flavoring, please click the Product details to view specification, application, pictures, prices and Other information about Food flavoring.
Whatever you are a group or individual, we will do our best to provide you with accurate and comprehensive message about Food flavoring!
Tartaric Acid, Vanillin, Menthol, Menthol Crystal, Flavoring agent, Baking powder
Xi'an Gawen Biotechnology Co., Ltd , https://www.ahualynbio.com