Shi Yigong first observed high-resolution human splice structure
On May 12, 2017, Shi Yigong, a research group of the Tsinghua University School of Life and the Center for Structural Biology and Innovation, published a book entitled "Atomic Resolution Structure of Human Splices" on Cell. Atomic Structure of the Human Spliceosome). This is the first high-resolution human splice structure, and it is the first time that a structure of a splice body from a higher organism other than yeast has been observed on a near-atomic resolution scale, further revealing the assembly and working mechanism of the splice. It provides an important basis for understanding the RNA splicing process of higher organisms.
In eukaryotic cells, most genes are discontinuous, and their coding regions are called "intron" non-coding sequence partitions. In the process of gene expression, the intron needs to be removed by a "scissing" and "splicing" chemical reaction, so that the coding region can be ligated into different messenger RNAs (mRNAs). The same gene, because of the different boundaries and numbers of introns, can be spliced ​​to produce a variety of mRNAs encoding proteins. RNA splicing is a unique process for all eukaryotes and one of the key steps in the “central rule†of eukaryotes. It is also considered to be an important molecular basis for the complexity of eukaryotes. The RNA splicing process must be highly accurate, and any error can lead to abnormal gene expression and even disease. According to statistics, more than 1 / 3 of hereditary diseases are associated with abnormal RNA splicing.

The complex physiological process of RNA splicing is performed by a spliceosome. The splice variant is the most complex macromolecular machine known in the cell. It has a large molecular weight and is highly dynamic. It is mainly composed of a protein-nucleic acid ribonucleoprotein (RNP) assembled on the precursor messenger RNA (pre-mRNA). Complete a series of processes such as identification, assembly, activation, catalysis, and finally dissociate into many small functional units after the reaction is completed to enter the next reaction cycle. According to the composition and conformation of the splice body, the splice body can be divided into several states such as E, A, B, Bact, B*, C, C*, P, and ILS (Fig. 1).
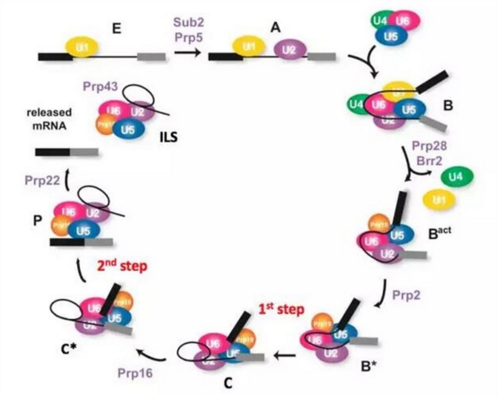
figure 1. Schematic diagram of the splicing reaction process (picture modified from Janine, 2012)
Analysis of the high resolution structure of the splice body is critical to understanding the mechanism of the splice reaction. In August 2015, Shi Yigong's research team took the lead in the world to analyze the high-resolution structure of yeast splicing bodies. In 2016, it also reported the high-resolution structure of yeast splicing bodies under different working conditions, providing the most Comprehensive and clear splice structure information reveals the molecular basis of RNA splicing and greatly advances the development of this field.
Canned Arbutus In Syrup,Arbutus In Syrup Tin,Arbutus In Syrup Canned,Arbutus Canned
ZHANGZHOU TAN CO. LTD. , https://www.zztancan.com