Yesterday, the star LncRNA hit the m6A and counterattacked today's new star.
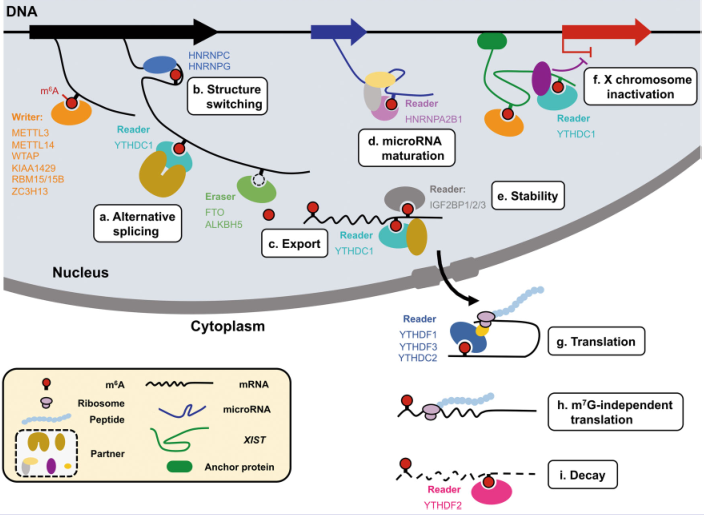
Tired of writer, eraser? Let's take a look at the reader. The YTHDF family is the most star-studded researcher in the reader due to its involvement in protein translation and degradation. It can be used in millions of m6A reader enzymes. The genes you study may be in addition to the YTHDF family. Other readers combined .

Impact factor: 7.8
Experimental methods: LncRNA sequencing , MeRIP-qPCR , RNA Pull Down , RIP , LncRNA quantitative PCR , mRNA quantitative PCR
Experimental materials: CRC clinical samples, etc., colorectal cancer cell lines, etc.
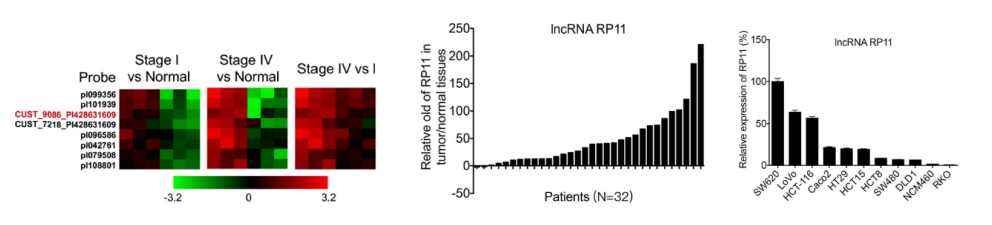
CRC tissues (cancer stage 1 and stage 3 cancer) and adjacent tissues were separately subjected to LncRNA sequencing (the cloud sequence can provide this service ) , and LncRNA-RP-RP11 which is highly differentially expressed in both stage 1 and stage 3 of cancer was screened. In the sequenced samples and 32 clinical sample tissues, it was confirmed by LncRNA quantitative PCR that it was highly expressed in the experimental group. The results of colon cancer and rectal cancer databases have also confirmed this trend of expression. RP11 is widely expressed in multiple colorectal cancer cell lines and is highly expressed in SW620 cells primary cultured in metastatic lymph node tissues of CRC patients.
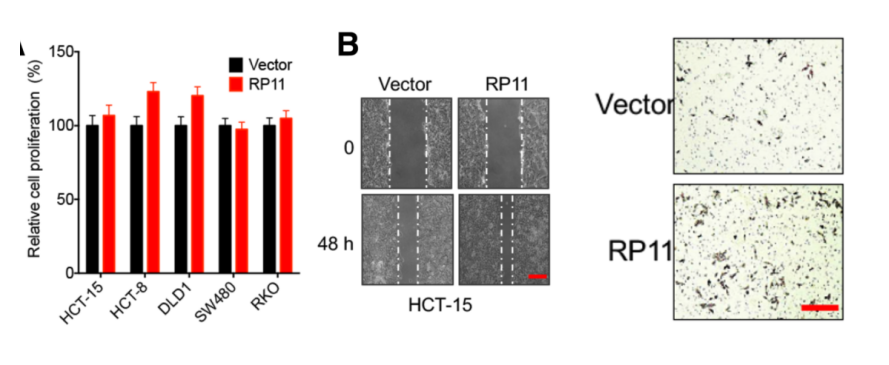
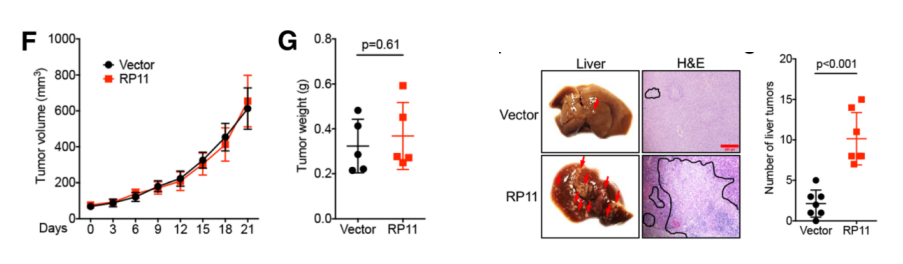
To further elucidate the functional mechanism of RP11 in vivo and in vitro. The authors first confirmed that the RP11 overexpressing cell line can significantly promote cell migration ability. Since this process affects the expression levels of marker proteins such as E-Cad, FN and Vim, it may be associated with EMT and cancer metastasis. Next, the authors highly expressed RP11 gene in xenografted nude mice and found that the gene can promote tumor proliferation. The authors introduced cancer cells stably expressing RP11 by tail injection in nude mice, and after 8 weeks of feeding, they were found to have metastasized to mouse liver tissues.
Studies have shown that LncRNA can influence the expression of transcripts around it through homeopathic regulation. Therefore, six transcripts were found around LncRNA RP11, namely NUDT12, C5orf30, PPIP5K2, GIN1, RP11-6 N13.1 and CTD-2374C24. It was confirmed by mRNA quantitative PCR that the above transcripts were not differentially expressed in the RP11 high expression group and the SW620 cell line with the highest RP11 expression. Therefore, the functional mechanism of RP11 is not affected by the mode of homeopathic regulation.
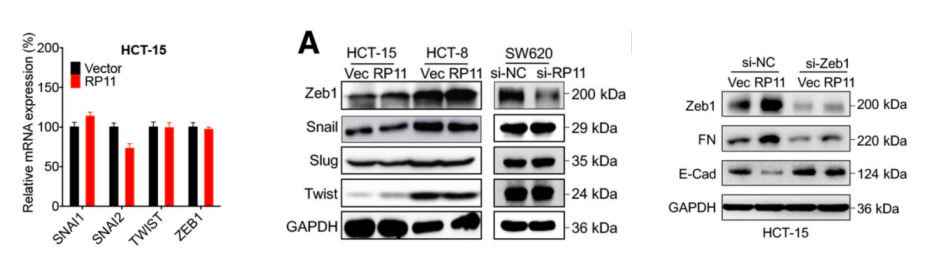
In order to confirm the above conjecture, the authors examined the expression levels of multiple EMT-related factors in mRNA over-expressed by over-expressing cells. The results showed that Zeb1 was positively correlated with the expression pattern of RP11. And in the Zeb1 overexpressing cell line, it was confirmed that the expression levels of the FN and Vim markers were affected. Therefore, the downstream of RP11 may be Zeb1.
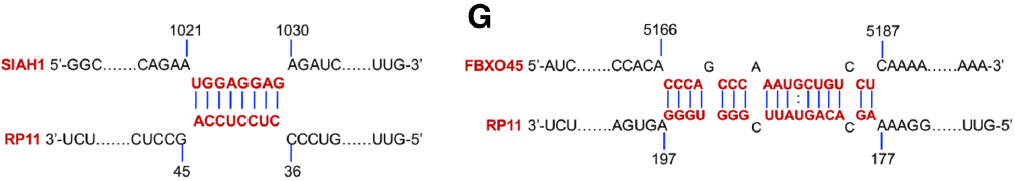
In order to verify the previous conjecture, the authors confirmed that Zeb1 and RP11 are not directly combined at the mRNA and protein levels by Zeb1 RIP-LncRNA PCR (the cloud sequence can provide this service) and RP11 Pull Down-mass spectrometry (the cloud sequence can provide this service) . Relationship.
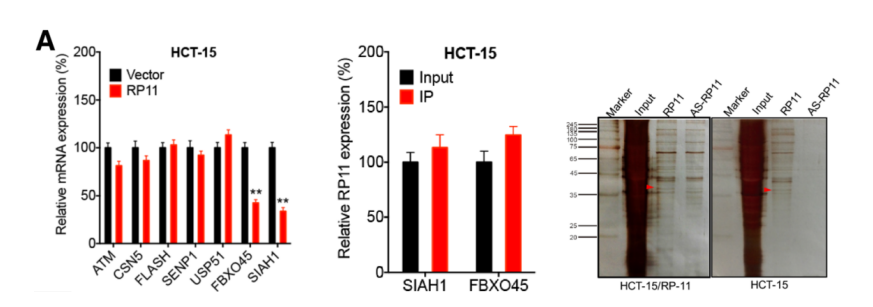
Similarly, the authors speculate that the downstream proteins may be Siah1 and Fbxo45. At the mRNA level, both of them regulated the expression of RP11 and Zeb1 by RP11 RIP-mRNA PCR . At the protein level, it was confirmed by RP11 Pull Down-mass spectrometry that the three were not directly bound.
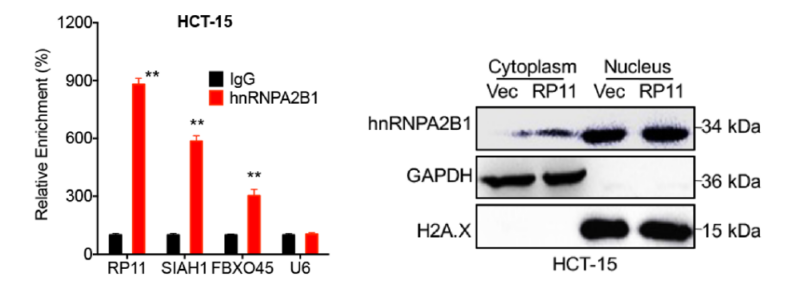
First, the authors suspected that high expression of RP11 was associated with DNA methylation. After treatment of cells with DNA methylation inhibitory enzymes, the expression of RP11 was found to be unchanged, confirming that RP11 expression in CRC cells was not associated with DNA methylation. At the same time, experiments have also confirmed that it is not related to histone acetylation.
Analysis of the expression of a large number of different stages of clinical samples showed that m6A modification first regulated RP11, and RP11 affected Siah1-Fbxo45/Zeb1. With the KM curve, the authors found that the survival of patients with high expression of RP11 rectal cancer is not optimistic. At the same time, the relationship between downstream genes and other clinical indicators of patients was also analyzed. Overall, it was confirmed that m6A/RP11/Zeb1 promoted the development of CRC.
In the present study, the authors first screened high-expression LncRNA-RP11 in the CRC disease group by LncRNA high-throughput sequencing and confirmed that it can promote tumor migration.
Subsequently, the authors aimed to investigate what is the downstream binding protein of RP11 and how it regulates the migration phenotype.
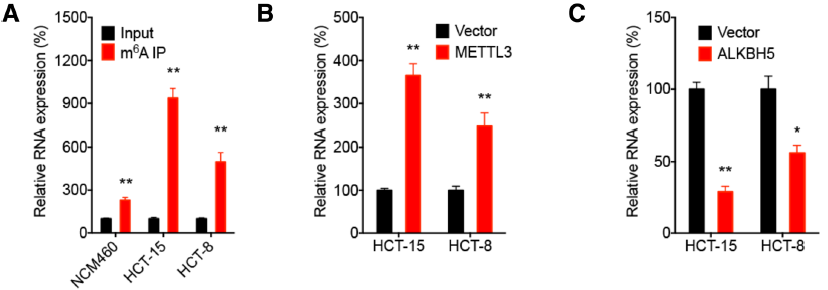
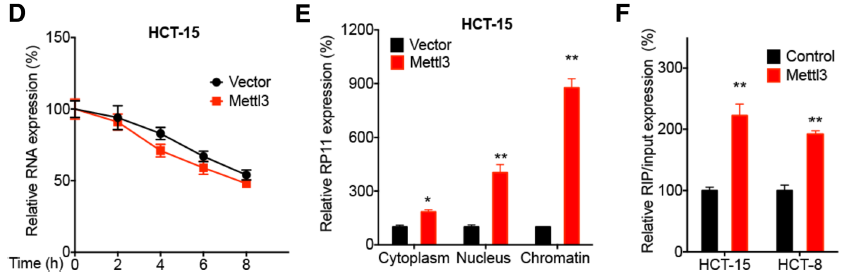
On the one hand: with LncRNA Pull Down , the protein that binds rapidly to RP11 is the m6A reader hnRNPA2B. By RIP-PCR , the RNA that quickly finds binding to this protein is RP11. On the other hand, by MeRIP-qPCR , it was confirmed that m6A modification was present on RP11 in the three disease-related cell lines. At the same time, Mettl3 promotes RP11 stability as its upstream, thereby promoting the development of CRC.

Cloud Biological Sequence provides a detection level of the overall methylation modification m6A colorimetric method, RNA methylation sequencing, MeRIP-qPCR verification, RIP, and RNA Pull Down Mechanism service. RNA methylation sequencing technology is the real implementation of m6A, m5C and m1A modification, detection of molecules in addition to mRNA, but also detection of circular RNA, LncRNA and other non-coding RNA. Since 2016, the number of samples has accumulated more than 5000+, and the integrated power of MeRIP has reached more than 98%.
Now, in order to solve the problem of small sample size of customers, ultra-micro-MeRIP sequencing technology is introduced, and 500 ng of total RNA can be used for sequencing experiments. Special samples can also be sequenced for RNA methylation, such as serum, plasma, exosomes, and paraffin samples.
Https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-019-1014-2
m6A full transcriptome sequencing
m6A LncRNA sequencing
Whole transcriptome sequencing
LncRNA sequencing
RNA Pull Down
RIP sequencing
Mass spectrometry
LncRNA quantitative PCR
Quantitative PCR
look here! How do miRNAs get high scores on m6A hotspots?
More than 10 points of m6A RNA methylation sequencing article----Cloud order bio-assisted
Occupy C! How does cloud-sequence bioanalysis quickly detect RNA methylation-related enzymes & RNA methylation levels in various cancers (on)
Occupy C! How does cloud-sequence bioanalysis quickly and simultaneously detect RNA methylation-related enzymes & RNA methylation levels in various cancers (2)
Cloud Sequence BioCustomer Latest Article: ChIP-seq & mRNA-seq Combined Application
Cloud order customer 12 points top article, teach you how to use RIP sequencing to play the molecular mechanism!
Nat. commun | Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis
Nature | SMAD2/3 and TGF-β pathway synergistically affect transcription factor m6A RNA methylation regulates stem cell development
Nature | m6A RNA methylation recognition protein YTHDF1 is involved in memory formation
No, Da Niu tells you how to study lncRNA methylation
Xiaobai must see! Summary of methods for identification of overall level of RNA methylation
Cloud Sequence Bio's Latest "RNA Methylation" Research Summary - Arabidopsis
Nature Breakthrough Research - RNA Methylation Newly Modified m1A Cloud Sequence Creature
The 2018 Nature Magazine's Breakthrough Research Results--m5C RNA Methylation Cloud Sequence Creature
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - non-coding RNA articles
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - virus articles
RNA methylation has already become like this, what are you waiting for! The latest cloud-order biological m6A "RNA methylation" research
Yunxu Bio exclusive M5C RNA methylation sequencing
RNA methylation shocked!
Shanghai Cloud-seq Biotech Co., Ltd.
Address: 3rd Floor, Building 20, No. 518, Banqiao Road, Songjiang District, Shanghai

Telephone Fax Website:
mailbox:
Double Head Ultrasound,Wireless Machine Double Head Linear,Double Head Probe Type Wireless Ultrasound,Double Head Wireless Ultrasound
Guangzhou Sonostar Technologies Co., Limited , https://www.sonoeye.com