The gospel of fat paper! Knocking out the Zfp217 gene can take away the "lifebuoy" in your waist through RNA m6A
Impact factor: 11.9
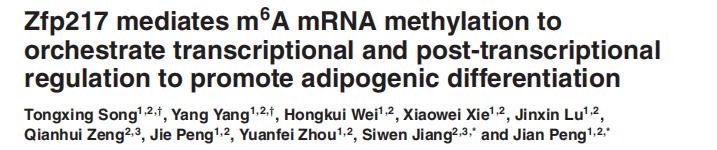
Experimental method: MeRIP-seq RNA-seq MeRIP-qPCR Co-IP ChIP-qPCR
Original link: Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation (doi: 10.1093/nar/gkz312)
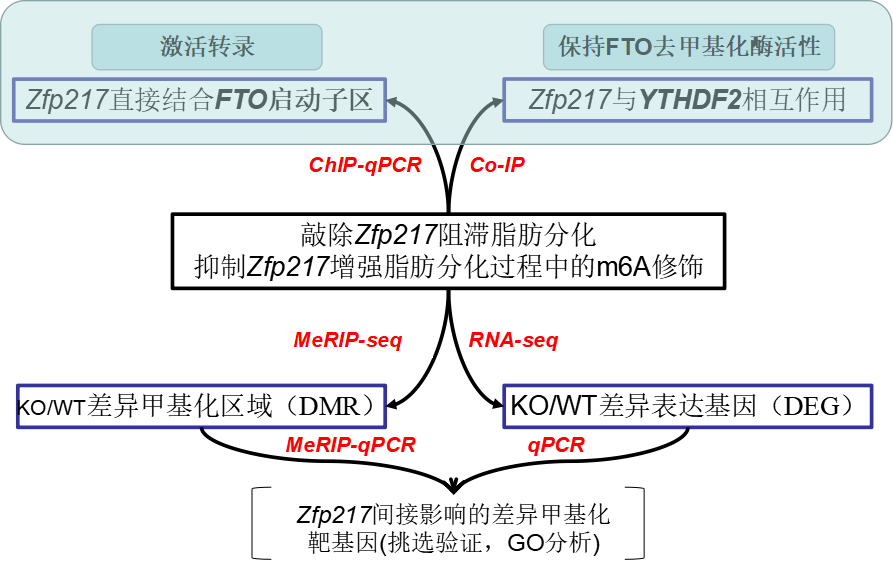
Figure 1: Zfp217 mediates mRNA m6A modification to regulate transcription and post-transcriptional modification to promote adipogenic differentiation
1. Zfp217 knockout significantly blocked fat differentiation
In 3T3L1 cells, both siRNA and CRISPR/Cas9 knockout Zfp217 showed significant blockade of fat formation, and the expression of related important genes such as PPARγ, P2, LPL, etc. were significantly down-regulated. The absence of Zpf217 clearly caused the formation of fat.
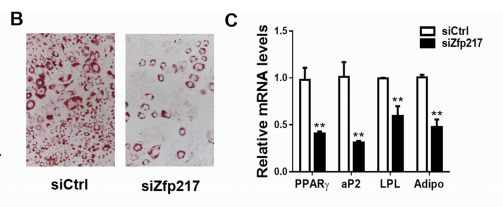
Figure 2: Significant down-regulation of adipose differentiation-related genes after Zfp217 knockout
The overall level of m6A modification (the cloud sequence provides this service) demonstrated that the m6A modification has a broad rise in Zfp217 knockout cells, and this increase occurs early in the knockout rather than 2 days after cell induction culture.
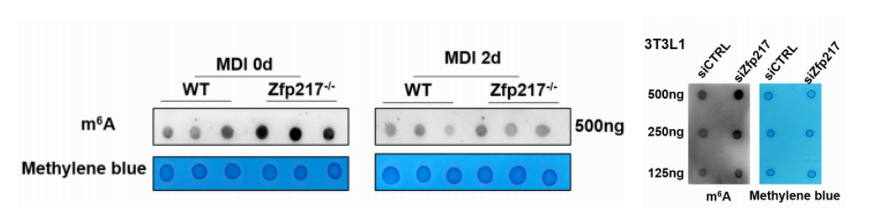
Figure 3: Overall level of mRNA m6A detection
RNA-seq sequencing was performed after Zfp217 knockout (the cloud sequence can provide this service) . Compared with wild type, 1431 genes were up-regulated and 941 genes were down-regulated. GO analysis indicated that most of the down-regulated genes and cell cycle, RNA processing As well as lipid biosynthesis, it is suggested that Zfp217 may be involved in liposome metabolism and RNA processing.
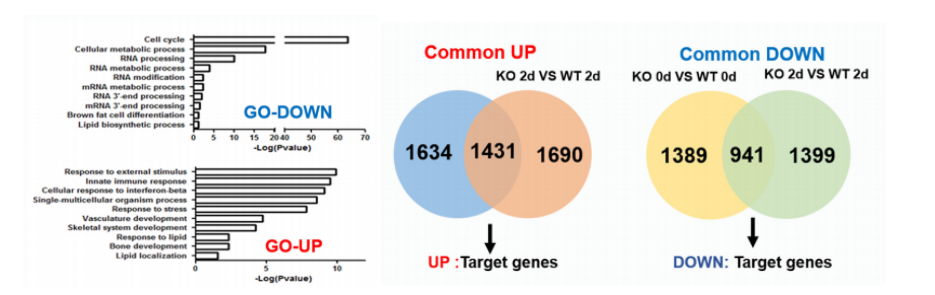
Figure 4: RNA-seq data collation and GO analysis results

Figure 5: MeRIP-seq RNA-seq combined analysis
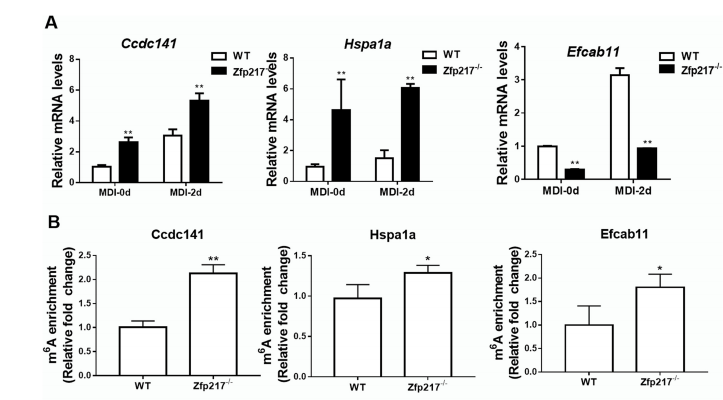
Figure 6: MeRIP-qPCR and qPCR verification
Studies in tumors have shown that Zfp217 can function in combination with other proteins. At this time, IP experiments with labeled overexpressing cell line HEK293T cells are disappointing results of Co-IP (the cloud sequence can provide this service) . The well-known FTO, ALKBH5 and METTL3/4 star methylases were not found, but in 3T3L1, the authors successfully demonstrated the binding of YTHDF2 and Zfp217 by Co-IP. This binding is independent of RNA. The results of cell localization indicated that both proteins were widely distributed outside the nuclear core, but Zfp217 was distributed more extranuclear than the intranuclear part. Laser confocal showed that the colocalization of YTHDF2 and Zfp217 indicated that they could synergistically play biological functions.
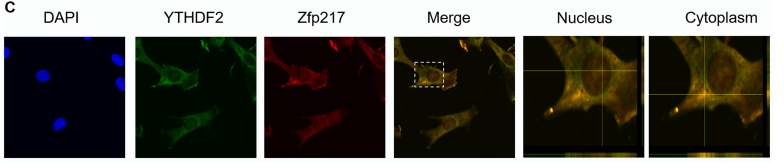
Figure 7: Subcellular localization shows that YTHDF2 and Zfp217 are co-localized in fine
5. Highlights: Zfp217 can directly act as a transcription factor to activate FTO transcription
Since Zfp217 has eight C2H2 zinc finger domains and a transfer activation domain, combined with biosignal analysis and ChIP-qPCR (the cloud sequence can provide this service) , the authors found that Zfp217 is indeed a transcription factor activity protein that can be combined with FTO. The promoter region regulates the transcriptional activity of the target gene, and FTO is one of the downstream target genes.
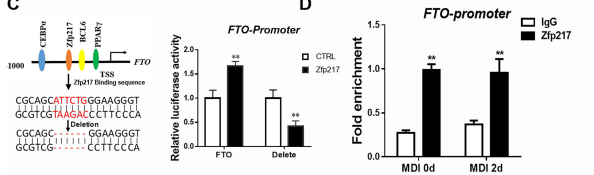
Figure 8: FTO TSS upstream combined with Zfp217
Cloud-sequence organisms provide colorimetric detection of overall m6A methylation modification levels, RNA methylation sequencing, MeRIP-qPCR validation, RIP and RNA Pull Down mechanisms . RNA methylation sequencing technology is the real implementation of m6A, m5C and m1A modification, detection of molecules in addition to mRNA, but also detection of circular RNA, LncRNA and other non-coding RNA. Since 2016, the number of samples has accumulated more than 5000+, and the integrated power of MeRIP has reached more than 98%.
Now, in order to solve the problem of small sample size of customers, ultra-micro-MeRIP sequencing technology is introduced, and 500 ng of total RNA can be used for sequencing experiments. Special samples can also be sequenced for RNA methylation, such as serum, plasma, exosomes, and paraffin samples.
m6A modified overall level detection
m6A full transcriptome sequencing
m6A LncRNA sequencing
Whole transcriptome sequencing
LncRNA sequencing
RNA Pull Down
RIP
Mass spectrometry
LncRNA quantitative PCR
Quantitative PCR
look here! How do miRNAs get high scores on m6A hotspots?
More than 10 points of m6A RNA methylation sequencing article----Cloud order bio-assisted
Occupy C! How does cloud-sequence bioanalysis quickly detect RNA methylation-related enzymes & RNA methylation levels in various cancers (on)
Occupy C! How does cloud-sequence bioanalysis quickly detect RNA methylation-related enzymes & RNA methylation in various cancers (2)
Cloud order customer 12 points top article, teach you how to use RIP sequencing to play the molecular mechanism!
Nat. commun | Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis
Nature | SMAD2/3 and TGF-β pathway synergistically affect transcription factor m6A RNA methylation regulates stem cell development
Nature | m6A RNA methylation recognition protein YTHDF1 is involved in memory formation
No, Da Niu tells you how to study lncRNA methylation
Xiaobai must see! Summary of methods for identification of overall level of RNA methylation
Cloud Sequence Bio's Latest "RNA Methylation" Research Summary - Arabidopsis
Nature Breakthrough Research - RNA Methylation Newly Modified m1A Cloud Sequence Creature
The 2018 Nature Magazine's Breakthrough Research Results--m5C RNA Methylation Cloud Sequence Creature
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - non-coding RNA articles
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - virus articles
RNA methylation has already become like this, what are you waiting for! The latest cloud-order biological m6A "RNA methylation" research
Yunxu Bio exclusive M5C RNA methylation sequencing
RNA methylation shocked!
Shanghai Yunxu Biological Technology Co., Ltd.
Shanghai Cloud-seq Biotech Co., Ltd.
Address: 3rd Floor, Building 20, No. 518, Banqiao Road, Songjiang District, Shanghai

Telephone Fax Website:
mailbox:
1. Material of our mildew tape
1) Transparent acrylic adhesive + polyester, thickness: 0.5mm and 0.8mm
2) PVC + acrylic adhesive, thickness is about 1.5mm, different colors for choosing.
3) PE + butyl adhesive, long serfice life and and good temperature resistance.
2. Features of our mildew tape
1) Good adhesion and soft material, can be adhered to many different surfaces.
2) Waterproof, resist the water, oil and dust go inside the cracks and corners, resist mildew.
3) Long service life, easy to clean.
4) Different colors and patterns for choosing.
5) Easy to operate, Do not need to use other complicated tools, the waterproof tape is affixed around the sink, use PE Self Adhesive, repeatedly press several times.It adhesive sticks well can seal the gap between the tile and the wall.
3. Applications of our mildew tape
Widely used for sealing the seams of gas stove, sink, basin, bathtub and walls, it could prevent them from getting mouldy and black, keep your kitchen and bathroom clean and new for a long time.
Mildew tape,Anti-mildew tape,anti mildew tape,caulking tape,caulking strip,caulk strip,caulk tape
Kunshan Jieyudeng Intelligent Technology Co., Ltd. , https://www.jerrytape.com