Mechanism of circRNA_104075 in promoting the occurrence and progression of liver cancer
CircRNA_104075 sponge adsorbs miR-582-3p, which inhibits YAP expression, thereby promoting the occurrence and progression of liver cancer
Article introduction:
Primary liver cancer is one of the top three causes of cancer-related deaths worldwide. Hepatocellular carcinoma (HCC) is the most common primary liver cancer. Due to the lack of high-specificity and sensitivity of early diagnostic biomarkers, HCC patients often do not receive timely and effective treatment. Compared to long-chain non-coding RNAs and miRNAs, circRNAs, as a novel circular RNA, have a covalently closed circular structure with highly stable expression and presence in tissues and blood. And studies have shown that circRNA plays a key regulatory role in the development of various diseases, and can be used as a biomarker with high specificity and sensitivity in early clinical diagnosis, and has a good application prospect. In addition, m6A RNA methylation modification is the most abundant type of RNA modification in mammals. It plays a variety of roles in the epigenetic regulation of RNA, including mRNA stability, alternative splicing, and miRNA maturation. m6A modifications can also modulate the structure of methylated transcripts, preventing or inducing binding of other RNAs.
Article content:
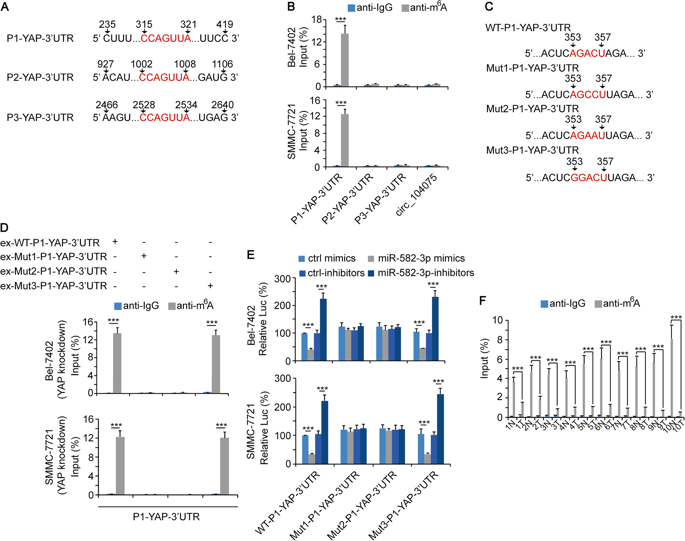
1. Circ_104075 is highly expressed in HCC tissues
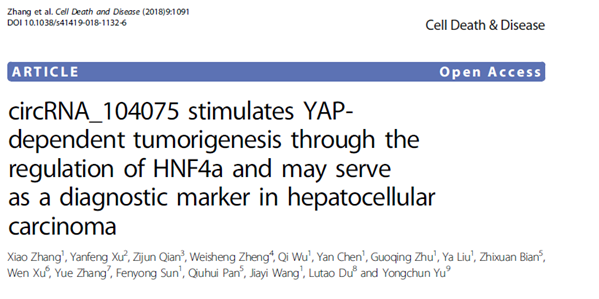
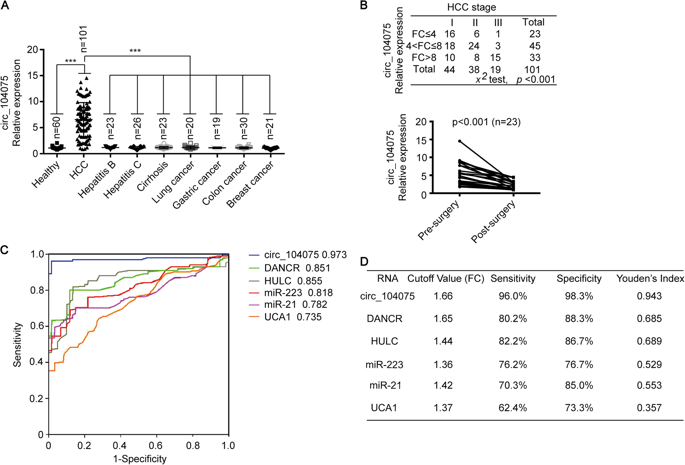
The team first combined the circRNA expression data (the cloud sequence organism provides circular RNA sequencing services ) in three previously reported HCC tissues, and screened a highly expressed circular RNA molecule, circ_104075. And compared with normal adjacent tissues, circ_104075 was significantly expressed in HCC. At the same time, compared with other HCC serodiagnostic markers (DANCR, HULC, UCA1, miR-21 and miR-223) reported in other previous studies, the level of circ_104075 expression was significantly higher in HCC patients.
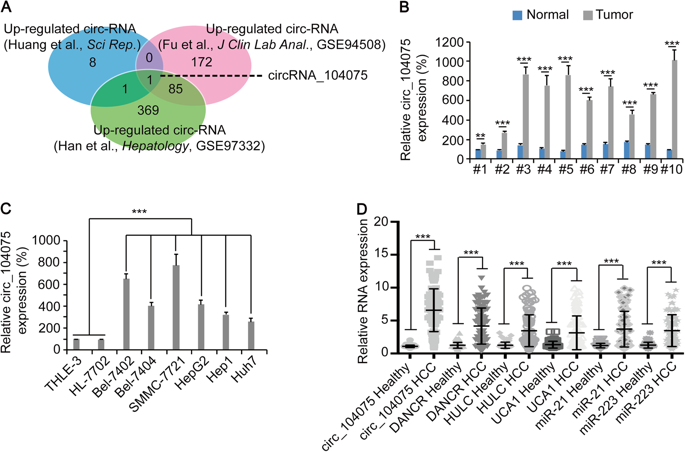
The authors selected a number of recognized HCC oncogenic transcription factors. By knockout and overexpression, only HNF4a expression was positively correlated with the expression of circ_104075. Compared with normal mice, the size and weight of liver in HNF4a knockout mice were significantly reduced, and the expression of circ_104075 was also significantly decreased. Clinical HCC tissue analysis revealed that HNF4a expression was also up-regulated in HCC patients, further confirming the expression of HNF4a. A conclusion that is positively related to the expression of circ_104075.
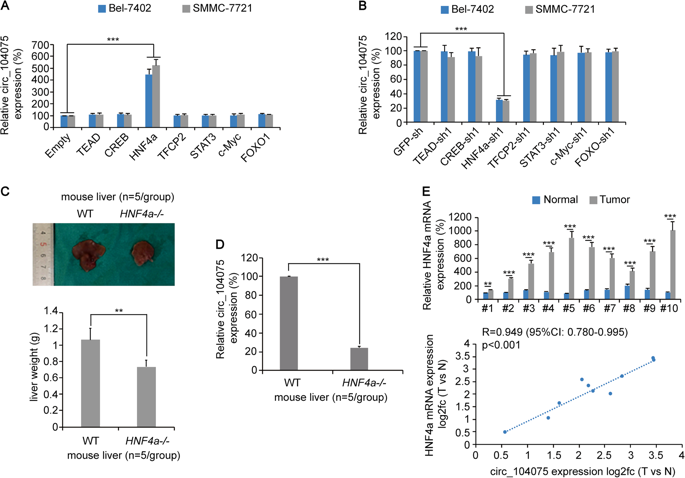
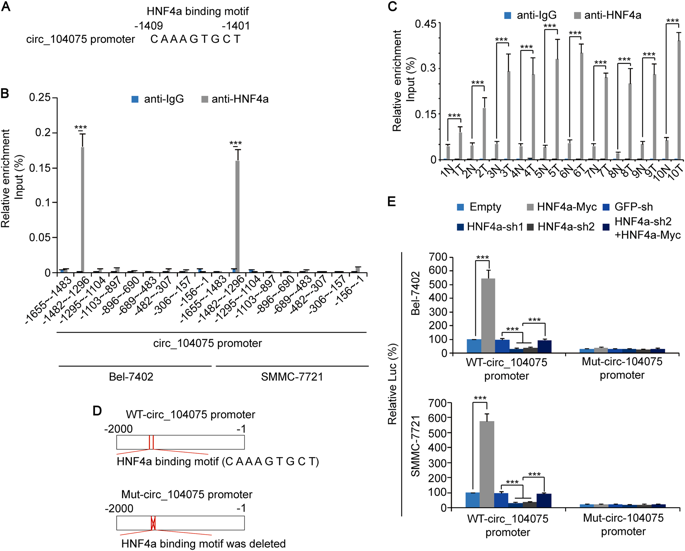
To verify how the transcription factor HNF4a regulates the expression of circ_104075, the authors predicted that HNF4a could bind to the site of the circ_104075 promoter (-1482 to -1296); then capture the HNF4a pellet using ChIP-qPCR (the cloud sequence organism provides this experiment ). After the complex, HNF4a was found to bind to the circ_104075 promoter (-1482 to -1296) in HCC tissues and HCC cell lines; luciferase reporter gene experiments also confirmed that overexpression of HNF4a can significantly promote the circ_104075 promoter. Luciferase activity, while knockdown of HNF4a expression inhibits luciferase activity of the circ_104075 promoter; if the HNF4a binding site is mutated on the circ_104075 promoter, overexpression or knockdown of HNF4a expression has no significant effect on luciferase activity, It was revealed that HNF4a positively regulates its expression by binding to the circ_104075 promoter.
Next, the authors explored which carcinogenic signaling pathways circ_104075 can regulate in HCC. By knocking out and over-expressing circ_104075, it was found that only YAP involved in the promotion of HCC signaling pathway changes, YAP was positively correlated with the expression of circ_104075, and YAP is a recognized HCC promoting factor. Clinical validation also revealed a positive correlation between the expression of YAP and the expression of circ_104075.
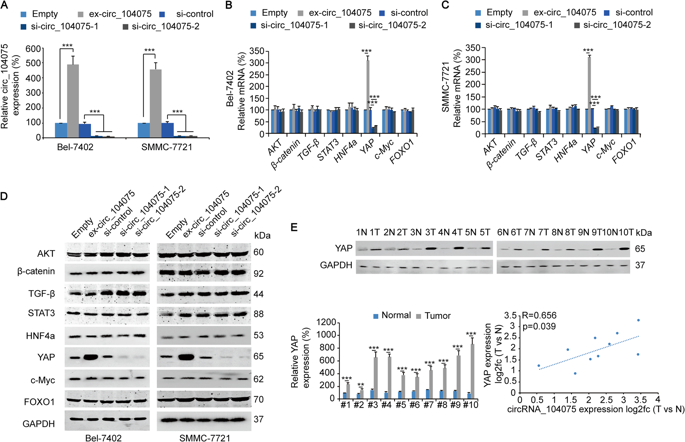
The authors first thought that circRNA adsorbs miRNA by sponge and directly regulates the expression of target protein. The authors used the miRanda database to predict the five miRNAs that circ_104075 are most likely to bind, and then capture the complex by RNA pull down technology (the cloud sequence organism provides this experiment ). RT-qPCR analysis revealed that only miR-582-3p was bound; by constructing the mutant forms circ_104075 and miR-582-3p, luciferase reporter gene experiments confirmed the binding of WT circ_104075 and WT miR-582-3p, WT miR -582-3p inhibited the enzymatic activity of WT-, Mut1- and Mut2-circ_104075, but did not inhibit the enzymatic activity of Mut3 circ_104075, suggesting that circ_104075 binds to the 5' and 3' ends of this binding site for miR-582-3p It is vital. At the same time, Mut miR-582-3p could not inhibit the enzymatic activity of circ_104075.
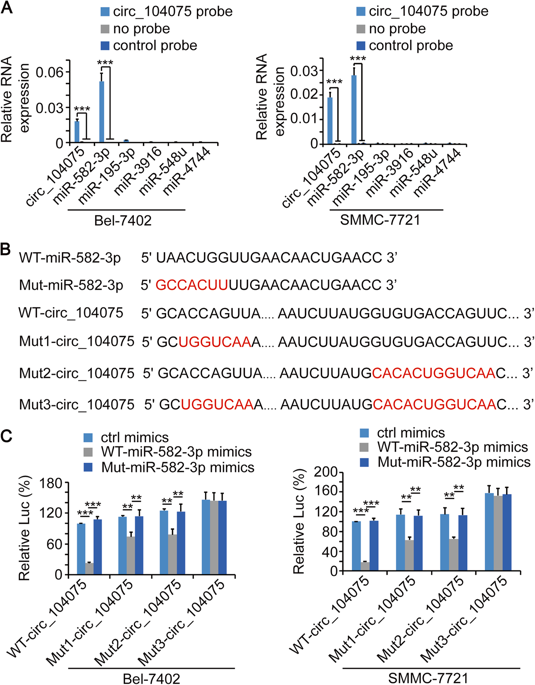
It has been confirmed that circ_104075 can bind to miR-582-3p, and circ_104075 is positively correlated with YAP expression. Then the question arises, does miR-582-3p target the regulation of YAP expression? Further studies confirmed a negative correlation between miR-582-3p expression and YAP expression, and it was predicted that the 5' end of miR-582-3p can bind to three sites of the 3'UTR of YAP mRNA, and these three sites After mutating and performing luciferase reporter gene experiments, it was found that miR-582-3p could not inhibit Mut1-YAP-3'UTR, indicating that the 3'UTR of YAP mRNA (315-321 base sequence) was 5 for miR-582-3p The binding of 'ends is important; by knocking down circ_104075 and inhibiting miR-582-3p, it was demonstrated that miR-582-3p plays an important role in promoting the expression of YAP in circ_104075.
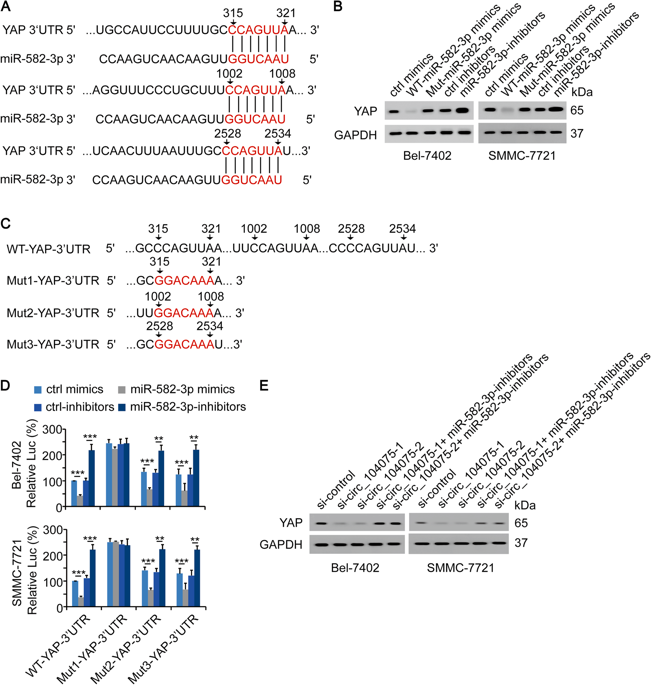
By MeRIP-qPCR (Biological Sequence cloud provide this experiment) detected P1-YAP-3'UTR (235-419 nucleotide sequence) upregulation, which region is also important for binding of miR-582-3p. In the 353-357 region of YAP-3'UTR (235-419 region), there is m6A motif (RRACU), AGACU, which was subjected to MeRIP-qPCR by mutating this point base and overexpressing, and found that only the last three bases were not mutated. The m6A modification level of the YAP-3'UTR of the base ACU is not lowered. At the same time, the luciferase reporter gene experiment also confirmed that the AGACU of the YAP-3'UTR 353-357 region is crucial for the inhibition function of miR-582-3p binding. MeRIP-qPCR of clinical tissue samples also confirmed that the level of m6A modification in the YAP-3'UTR 353-357 region of HCC patients was significantly reduced, consistent with the up-regulation of YAP expression in HCC patients.
Through analysis of a series of digestive diseases, it is found that circ_104075 is highly specific in the serum of HCC patients compared with other digestive system cancers, and is positively correlated with the progression of HCC, and is also in the serum of HCC patients after surgery. The expression level of circ_104075 was significantly reduced; ROC curve analysis found that serum circ_104075 expression level has higher sensitivity and specificity than other biomarkers in HCC, suggesting that it can be used as a biomarker for the diagnosis of HCC. It is thus used in the clinic ( cloud sequence organisms provide circular RNA real-time quantitative PCR services ).
Full text link
Https://
Cloud order related product recommendation
m6A RNA whole transcriptome methylation sequencing
m5C RNA whole transcriptome methylation sequencing
m1A RNA whole transcriptome methylation sequencing
Ultra-micro-m6A methylation sequencing colorimetric assay for overall methylation level transcriptome sequencing circular RNA sequencing
ChIP sequencing
RNA pull down
RIP sequencing
Past review
Nature | SMAD2/3 and TGF-β pathway synergistically affect transcription factor m6A RNA methylation regulates stem cell development
Nat. commun|Mettl3-mediated m6A RNA methylation regulates bone marrow mesenchymal stem cells and osteoporosis
Nature|m6A RNA methylation recognition protein YTHDF1 is involved in memory formation
No, Da Niu tells you how to study lncRNA methylation
RNA methylation enters the ultra-micro era
More than 10 points of m6A RNA methylation sequencing article--Cloud order bio-assisted
RNA methylation in 20 points, coexistence of heat and strength
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - non-coding RNA articles
Cloud Sequence Bio's Latest "RNA Methylation" Research Summary - Arabidopsis
Cloud Sequence Bio's latest m6A "RNA methylation" research summary - virus articles
Plant Cell: Arabidopsis found a new m6A RNA demethylation modifying enzyme
Cloud order customer 12 points top article, teach you how to use RIP sequencing to play the molecular mechanism!
2018 Nature Magazine's Breakthrough Research Results--m5C RNA Methylation Cloud Sequence Creature
Nature Breakthrough Research - RNA Methylation Newly Modified m1A Cloud Sequence Creature
Nature Heavy: m6A RNA methylation is involved in the key aspects of hematopoietic stem cell development!
Cancer Cell: Professor He Chuan discovers important functions of m6A RNA methylation
Xiaobai must see! Summary of methods for identification of overall level of RNA methylation
Hepatology: m6A RNA methylase METTL3 promotes the development of liver cancer
Yunxu Bio exclusive m5C RNA methylation sequencing
Nature article reveals multiple functions of RNA methylation
Filling the gap of epigenetic modification: a new mechanism for RNA methylation regulation gene export
Shanghai Yunxu Biological Technology Co., Ltd.
Shanghai Cloud-seq Biotech Co., Ltd.
Address: 3rd Floor, Building 20, No. 518, Zhangzhu Road, Songjiang District, Shanghai 
Telephone Fax Website:
mailbox:
Tetanus Toxoid Vaccine,Toxoid Vaccine,Hep B Immune Globulin,Immunoglobulin Injections
FOSHAN PHARMA CO., LTD. , https://www.fospharma.com